Dataframes (Pandas) and Plotting (Matplotlib/Seaborn)
Written by Jin Cheong & Luke Chang with modifications and additional by Todd Gureckis
In this lab we are going to learn how to load and manipulate datasets in a dataframe format using Pandas and create beautiful plots using Matplotlib and Seaborn. Pandas is akin to a data frame in R and provides an intuitive way to interact with data in a 2D data frame. Matplotlib is a standard plotting library that is similar in functionality to Matlab's object oriented plotting. Seaborn is also a plotting library built on the Matplotlib framework which carries useful pre-configured plotting schemes.
After the tutorial you will have the chance to apply the methods to a new set of data.
Also, here is a great set of notebooks that also covers the topic. In addition, here is a brief video providing a useful introduction to pandas.
First, we load the basic packages we will be using in this tutorial. Notice how we import the modules using an abbreviated name. This is to reduce the amount of text we type when we use the functions.
Note: %matplotlib inline is an example of 'cell magic' and enables plotting within the notebook and not opening a separate window. In addition, you may want to try using %matplotlib notebook, which will allow more interactive plotting.
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
Pandas has many ways to read data different data formats into a dataframe. Here we will use the pd.read_csv function.
df = pd.read_csv('http://gureckislab.org/courses/fall19/labincp/data/salary.csv', sep = ',', header='infer')
You can always use the ? to access the docstring for a function for more information about the inputs and general useage guidelines.
pd.read_csv?
df
However, often the dataframes can be large and we may be only interested in seeing the first few rows. df.head() is useful for this purpose. shape is another useful method for getting the dimensions of the matrix. We will print the number of rows and columns in this data set by using output formatting. Use the % sign to indicate the type of data (e.g., %i=integer, %d=float, %s=string), then use the % followed by a tuple of the values you would like to insert into the text. See here for more info about formatting text.
print('There are %i rows and %i columns in this data set' % df.shape)
df.head()
On the top row, you have column names, that can be called like a dictionary (a dataframe can be essentially thought of as a dictionary with column names as the keys). The left most column (0,1,2,3,4...) is called the index of the dataframe. The default index is sequential integers, but it can be set to anything as long as each row is unique (e.g., subject IDs)
print("Indexes")
print(df.index)
print("Columns")
print(df.columns)
print("Columns are like keys of a dictionary")
print(df.keys())
You can access the values of a column by calling it directly. Double bracket returns a dataframe
df[['salary']]
Single bracket returns a Series
df['salary']
You can also call a column like an attribute if the column name is a string
df.salary
You can create new columns to fit your needs. For instance you can set initialize a new column with zeros.
df['pubperyear'] = 0
Here we can create a new column pubperyear, which is the ratio of the number of papers published per year
df['pubperyear'] = df['publications']/df['years']
df.head()
df.loc[0, ['salary']]
Next we wil try .iloc. This method references the implicit python index (starting from 0, exclusive of last number). You can think of this like row by column indexing using integers.
df.iloc[0:3, 0:3]
Let's make a new data frame with just Males and another for just Females. Notice, how we added the .reset_index(drop=True) method? This is because assigning a new dataframe based on indexing another dataframe will retain the original index. We need to explicitly tell pandas to reset the index if we want it to start from zero.
male_df = df[df.gender == 0].reset_index(drop=True)
female_df = df[df.gender == 1].reset_index(drop=True)
Boolean or logical indexing is useful if you need to sort the data based on some True or False value.
For instance, who are the people with salaries greater than 90K but lower than 100K ?
df[ (df.salary > 90000) & (df.salary < 100000)]
Dealing with missing values
It is easy to quickly count the number of missing values for each column in the dataset using the isnull() method. One thing that is nice about Python is that you can chain commands, which means that the output of one method can be the input into the next method. This allows us to write intuitive and concise code. Notice how we take the sum() of all of the null cases.
The isnull() method will return a dataframe with True/False values on whether a datapoint is null or not a number (nan).
df.isnull()
We can chain the .null() and .sum() methods to see how many null values are added up.
df.isnull().sum()
You can use the boolean indexing once again to see the datapoints that have missing values. We chained the method .any() which will check if there are any True values for a given axis. Axis=0 indicates rows, while Axis=1 indicates columns. So here we are creating a boolean index for row where any column has a missing value.
df[df.isnull().any(axis=1)]
You may look at where the values are not null. Note that indexes 18, and 24 are missing.
df[~df.isnull().any(axis=1)]
There are different techniques for dealing with missing data. An easy one is to simply remove rows that have any missing values using the dropna() method.
df = df.dropna()
Now we can check to make sure the missing rows are removed. Let's also check the new dimensions of the dataframe.
print('There are %i rows and %i columns in this data set' % df.shape)
df.isnull().sum()
df.describe().transpose()
We can also get quick summary of a pandas series, or specific column of a pandas dataframe.
df.departm.describe()
Manipulating data in Groups
One manipulation we often do is look at variables in groups.
One way to do this is to usethe .groupby(key) method.
The key is a column that is used to group the variables together.
For instance, if we want to group the data by gender and get group means, we perform the following.
df.groupby('gender').mean()
Other default aggregation methods include .count(), .mean(), .median(), .min(), .max(), .std(), .var(), and .sum()
Before we move on, it looks like there were more than 2 genders specified in our data. This is likely an error in the data collection process so let recap on how we might remove this datapoint.
df[df['gender']==2]
replace original dataframe without the miscoded data
df = df[df['gender']!=2]
Now we have a corrected dataframe!
df.groupby('gender').mean()
Another powerful tool in Pandas is the split-apply-combine method.
For instance, let's say we also want to look at how much each professor is earning in respect to the department.
Let's say we want to subtract the departmental mean from professor and divide it by the departmental standard deviation.
We can do this by using the groupby(key) method chained with the .transform(function) method.
It will group the dataframe by the key column, perform the "function" transformation of the data and return data in same format.
To learn more, see link here
# key: We use the departm as the grouping factor.
key = df['departm']
# Let's create an anonmyous function for calculating zscores using lambda:
# We want to standardize salary for each department.
zscore = lambda x: (x - x.mean()) / x.std()
# Now let's calculate zscores separately within each department
transformed = df.groupby(key).transform(zscore)
df['salary_in_departm'] = transformed['salary']
Now we have salary_in_departm column showing standardized salary per department.
df.head()
pd.concat([femaledf, maledf],axis = 0)
We can reset the index to start at zero using the .reset_index() method
pd.concat([maledf, femaledf], axis = 0).reset_index(drop=True)
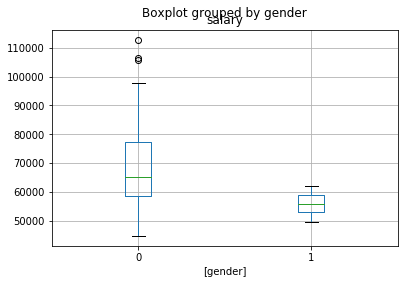
df[['salary','gender']].boxplot(by='gender')
df[['salary', 'years']].plot(kind='scatter', x='years', y='salary')
Plotting Categorical Variables. Replacing variables with .map
If we want to plot department on the x-axis, Pandas plotting functions won't know what to do because they don't know where to put bio or chem on a numerical x-axis. Therefore one needs to change them to numerical variable to plot them with basic functionalities (we will later see how Seaborn sovles this).
df['dept_num'] = 0
df.loc[:, ['dept_num']] = df.departm.map({'bio':0, 'chem':1, 'geol':2, 'neuro':3, 'stat':4, 'physics':5, 'math':6})
df.tail()
Now plot all four categories
f, axs = plt.subplots(1, 4, sharey=True)
f.suptitle('Salary in relation to other variables')
df.plot(kind='scatter', x='gender', y='salary', ax=axs[0], figsize=(15, 4))
df.plot(kind='scatter', x='dept_num', y='salary', ax=axs[1])
df.plot(kind='scatter', x='years', y='salary', ax=axs[2])
df.plot(kind='scatter', x='age', y='salary', ax=axs[3])
The problem is that it treats department as a continuous variable.
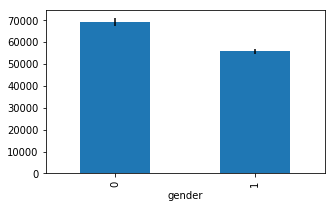
means = df.groupby('gender').mean()['salary']
errors = df.groupby('gender').std()['salary'] / np.sqrt(df.groupby('gender').count()['salary'])
ax = means.plot.bar(yerr=errors,figsize=(5,3))
Matplotlib
Matplotlib is an object oriented plotting library in Python that is loosely based off of plotting in matlab. It is the primary library that many other packages build on. Here is a very concise and helpful introduction to plotting in Python. Learn other matplotlib tutorials here
create a basic lineplot
plt.figure(figsize=(8, 4))
plt.plot(range(0, 10),np.sqrt(range(0,10)), linewidth=3)
plt.figure(figsize=(4, 4))
plt.scatter(df.salary, df.age, color='b', marker='*')
subplots allows you to control different aspects of multiple plots
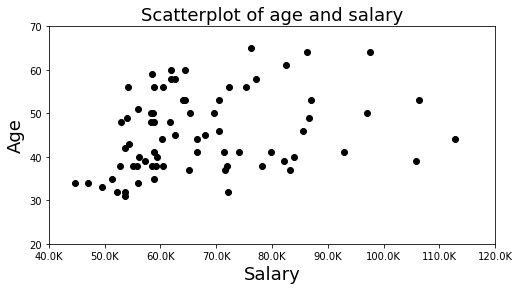
f,ax = plt.subplots(nrows=1, ncols=1, figsize=(8, 4))
ax.scatter(df.salary, df.age,color='k', marker='o')
ax.set_xlim([40000,120000])
ax.set_ylim([20,70])
ax.set_xticklabels([str(int(tick)/1000)+'K' for tick in ax.get_xticks()])
ax.set_xlabel('Salary', fontsize=18)
ax.set_ylabel('Age', fontsize=18)
ax.set_title('Scatterplot of age and salary', fontsize=18)
We can save the plot using the savefig command.
f.savefig('MyFirstPlot.png')
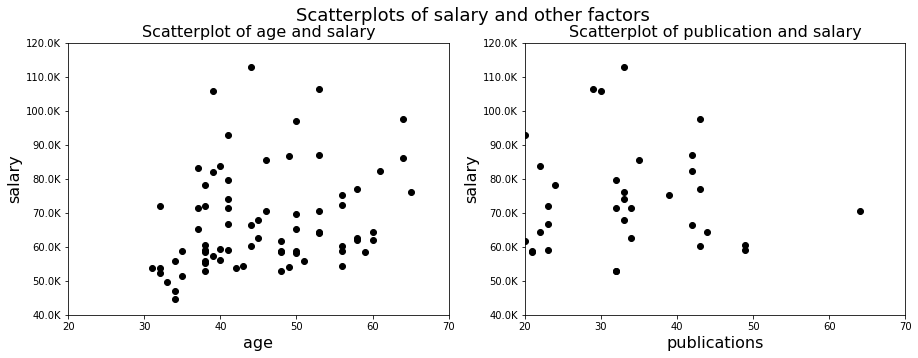
f,axs = plt.subplots(1, 2, figsize=(15,5))
axs[0].scatter(df.age, df.salary, color='k', marker='o')
axs[0].set_ylim([40000, 120000])
axs[0].set_xlim([20, 70])
axs[0].set_yticklabels([str(int(tick)/1000)+'K' for tick in axs[0].get_yticks()])
axs[0].set_ylabel('salary', fontsize=16)
axs[0].set_xlabel('age', fontsize=16)
axs[0].set_title('Scatterplot of age and salary', fontsize=16)
axs[1].scatter(df.publications, df.salary,color='k',marker='o')
axs[1].set_ylim([40000, 120000])
axs[1].set_xlim([20, 70])
axs[1].set_yticklabels([str(int(tick)/1000)+'K' for tick in axs[1].get_yticks()])
axs[1].set_ylabel('salary', fontsize=16)
axs[1].set_xlabel('publications', fontsize=16)
axs[1].set_title('Scatterplot of publication and salary', fontsize=16)
f.suptitle('Scatterplots of salary and other factors', fontsize=18)
Seaborn
Seaborn is a plotting library built on Matplotlib that has many pre-configured plots that are often used for visualization. Other great tutorials about seaborn are here
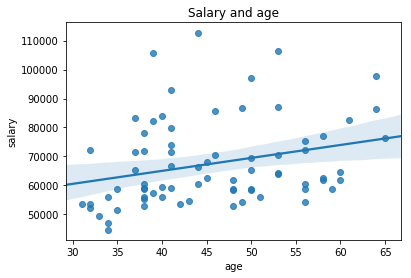
ax = sns.regplot(df.age, df.salary)
ax.set_title('Salary and age')
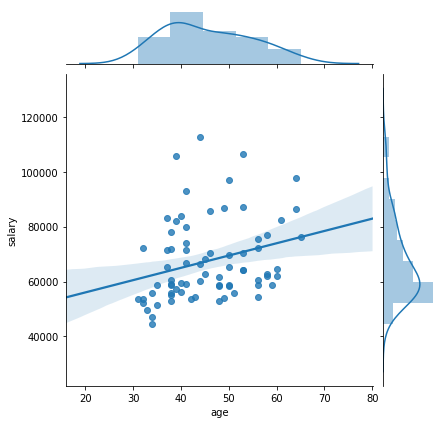
sns.jointplot("age", "salary", data=df, kind='reg')
sns.catplot(x='departm', y='salary', hue='gender', data=df, ci=68, kind='bar')
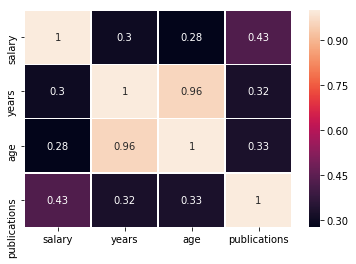
sns.heatmap(df[['salary', 'years', 'age', 'publications']].corr(), annot=True, linewidths=.5)